Genetic algorithms with R¶
Created by Ramses Alexander Coraspe Valdez¶
Created on July 1, 2020¶
Looking for extreme of a function/GA tries to find minimum of the function¶
[Start] Generate random population of n chromosomes (suitable solutions for the problem)
[Fitness] Evaluate the fitness f(x) of each chromosome x in the population
[New population] Create a new population by repeating following steps until the new population is complete
[Selection] Select two parent chromosomes from a population according to their fitness (the better fitness, the bigger chance to be selected)
[Crossover] With a crossover probability cross over the parents to form a new offspring (children). If no crossover was performed, offspring is an exact copy of parents
[Mutation] With a mutation probability mutate new offspring at each locus (position in chromosome).
[Accepting] Place new offspring in a new population
[Replace] Use new generated population for a further run of algorithm
[Test] If the end condition is satisfied, stop, and return the best solution in current population
[Loop] Go to step 2
Installing libraries¶
install.packages("genalg")
library(genalg) #A R based genetic algorithm that optimizes, using a user set evaluation function, a binary chromosome which can be used for variable selection. The optimum is the chromosome for which the evaluation value is minimal.
Importing libraries¶
library(genalg)
library(parallel)
library(ggplot2)
detectCores(all.tests = FALSE, logical = TRUE)
Knapsack problem¶
weight.limit <- 10
items <- data.frame(
item=c("encendedor","casa_camp","navaja","linterna",
"manta","sleep_bag","brujula", "agua.5.litro",
"atun.kilo", "cuchillo", "cerillos", "bat.extra"
,"pedernal", "jabon", "cepillo", "papel.higie",
"barritas.avena","kit.pesca", "cazuela", "botiquin",
"carne.seca","cerveza", "mezcal", "hielos",
"bloqueador_solar","camisa_larga","sombrero","gps",
"paneles_solares", "mapa",
"repelente", "zapatos especiales", "cuerdas",
"hacha", "escopeta", "telefono", "balas"),
survivalpoints = c(90,95,85,70,
50,60,80,100,
80,85,70,50,
80,40,40,10,
40,5,55,99,
99,5,5,2,
5,80,50,90,
50,99,
20, 50, 60,
70, 60, 20, 50),
weight = c(.01, 5,.1,.5,
2,1,.01, 5,
1,.1,.01,.5,
1,.1,.01,.1,
.5,2,.5,.5,
1,1,1,1,
.5,.2,.3,1,
2,.3,
.3, .5, 1,
1, 1, 0.4, 0.6)
)
items
fitness.generic <- function(x) {
items.weight <- x %*% items$weight
items.s.p <- x %*% items$survivalpoints
if (items.weight > weight.limit)
{
return(0)
}
else
{
return (-items.s.p)
}
}
?rbga.bin
ga.one <- rbga.bin(size=37,
popSize=200,
iters=200,
mutationChance=0.01,
elitism = 4,
evalFunc = fitness.generic,
verbose = T
)
best <- ga.one$population[ga.one$evaluations == min(ga.one$best),][1,]
best.items <- items$item[best == 1]
best.items
Finalweights <- best %*% items$weight
Finalweights
Finalsurvivalpoints <- best %*% items$survivalpoints
Finalsurvivalpoints
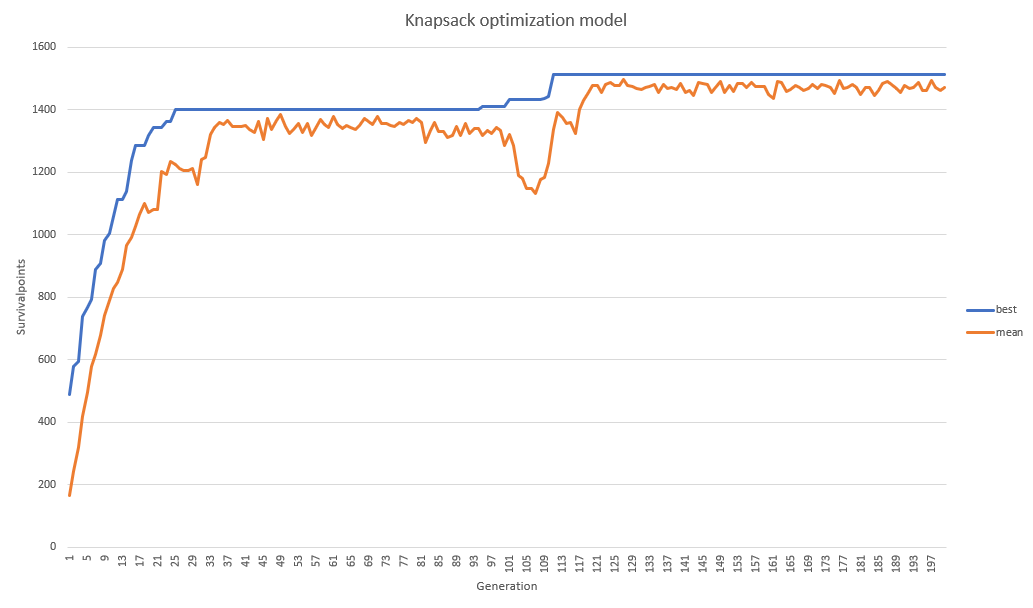
The above result shows that the items in the backpack should be: encendedor,navaja,linterna,brujula,atun.kilo,cuchillo,cerillos,pedernal,jabon,cepillo,barritas.avena,cazuela,botiquin,carne.seca,camisa_larga,gps,mapa,zapatos especiales,cuerdas,hacha,balas with a total weight of 9.94 kilos, which meets the limit of 10 kilos, and a survivalpoint of 1512. however, the choice of a "linterna" without "bat.extra" should be punished, now, let's improve the fittnes function to give more information to the genetic algorithm, because the presence of some elements are unnecessary if they are not supported by the presence of others.¶
Improving Fitness Function¶
fit.punish <- function(x){
punishment <- 0
#"linterna vs bat.extra" , "escopeta vs balas"
p <- list('1'=c(4,12), '2'=c(35,37))
for (v in p) {
result <- x[v[1]] - x[v[2]]
if(result < 0 ){
punishment <- punishment + (items$survivalpoints[v[1]] + items$survivalpoints[v[2]])
}
else if(result > 0) {
punishment <- punishment + items$survivalpoints[v[1]]
}
else {
}
}
return (punishment)
}
fitness.generic.2 <- function(x) {
items.weight <- x %*% items$weight
items.s.p <- x %*% items$survivalpoints
if (items.weight > weight.limit){
return(0)
}
else{
fit.val <- items.s.p - fit.punish(x)
return (-fit.val)
}
}
?rbga.bin
ga.one <- rbga.bin(size=37,
popSize=200,
iters=200,
mutationChance=0.01,
elitism = 4,
evalFunc = fitness.generic.2,
verbose = T
)
best <- ga.one$population[ga.one$evaluations == min(ga.one$best),][1,]
best.items <- items$item[best == 1]
best.items
Finalweights <- best %*% items$weight
Finalweights
Finalsurvivalpoints <- best %*% items$survivalpoints
Finalsurvivalpoints
Conclusions¶
The improved fitness function offers correct results, the presence or absence of elements in the Knapsack that depend on others is being punished, therefore the function will always try to include both. "linterna, bat.extra" and "escopeta , balas"